Network Modelling for Aquatic Animal Disease
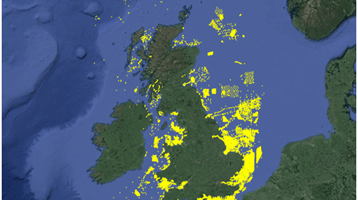
Network models have been applied to investigate disease spread in terrestrial animal systems, but not some widely applied or aquatic animal diseases, to support the development of policies to control disease outbreaks. We have developed spatially explicit, stochastic, state-transition (SIR) model of trout farms in England and Wales (E&W) and used social network analysis (SNA) to examine connectivity of fish farms in both salmonid and cyprinid farm networks. The SIR model used farm level fish movement and other data to quantify the following routes of pathogen spread between farms: live animal movements, river contact, mechanical (i.e. fomites). The network was used to simulate the spread of viral haemorrhagic septicaemia (VHS). The key findings were that the spread of the virus could be significantly impacted by the restricting movement of live fish from highly connected sites. The importance of contact chasing and disease detection rates were highlighted. The model allows for ‘what if’ questions to be asked to refine current control strategies.
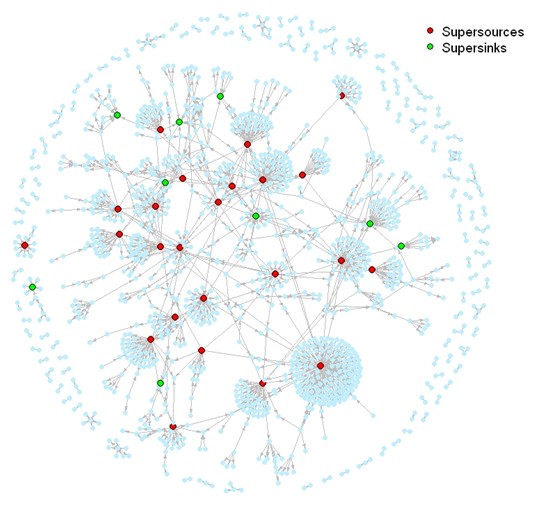
Social network analysis (SNA) models indicated potential for more frequent yet smaller scale disease outbreaks in the cyprinid industry and less frequent but larger scale outbreaks in the salmonid industry. Each dot in the image represents a node, and each grey line a contact between nodes. The super-sources (red nodes) are those that supply fish to at least 10 other sites, while the super-sinks (green nodes) receive fish from at least five. High connectivity between river catchments within both networks was shown, posing challenges for zoning at the catchment level for the purpose of disease management. The models can be used to inform both routine risk based surveillance of fish farms and surveys undertaken in the face of an outbreak.
Related to this article
Case studies
People
Further Reading

Working for a sustainable blue future
Our Science